Madrid Pollution
All the files of this project are saved in a GitHub repository.
Dataset of the Pollution Level in Madrid
This report describes an analysis of the pollution in Madrid between 2011 and 2016.
The dataset consists in:
- 72 csv files containing hourly measures of pollutants across 24 stations,
- 1 xlsx file containing daily weather information.
The stations are located all across the city:
Packages
This analysis requires these R packages:
- Data Cleaning:
readxl,tidyr - Plotting:
ggplot2,corrplot,GGally,gridExtra,leaflet - Statistics:
jtools,lattice,car,caret,MASS
These packages are installed and loaded if necessary by the main script.
Data Preparation
The pollution and weather data are first read from the input files, formatted, combined and aggregated, into the data frame pollution_daily_h which provides the averaged information per day.
The dataset contains information for 12 pollutants: NO2, SO2, O3, PM2.5, BEN, CO, EBE, NMHC, NO, PM10, TCH, TOL.
The workflow to prepare the data is as below:
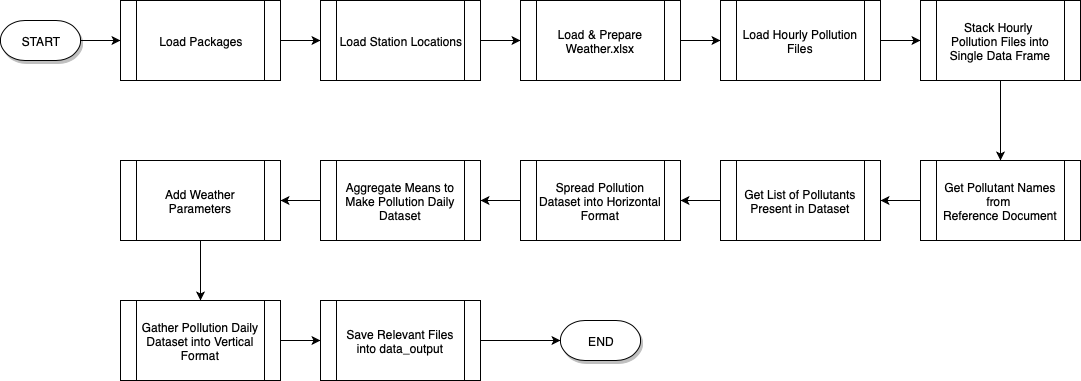
Additional variables have been added:
- month: the first day of the related month
- week: the first day of the related week
- temp_gap: difference between temp_min and temp_max
The data frame pollution_daily_h is structured as below:
str(pollution_daily_h)
## 'data.frame': 2192 obs. of 22 variables:
## $ date : Date, format: "2011-01-01" "2011-01-02" ...
## $ NO2 : num 41.5 48.5 63.6 46.3 51.5 ...
## $ SO2 : num 10.71 11.93 11.91 8.84 9.51 ...
## $ O3 : num 20.47 15.56 9.45 13.34 10.88 ...
## $ PM2.5 : num 9.36 9.08 11.94 9.4 10.51 ...
## $ BEN : num 0.765 0.869 1.176 0.857 0.856 ...
## $ CO : num 0.372 0.453 0.532 0.367 0.395 ...
## $ EBE : num 0.595 0.722 1.047 0.788 1.074 ...
## $ NMHC : num 0.187 0.208 0.246 0.196 0.196 ...
## $ NO : num 16.5 28.8 71.1 27.8 36 ...
## $ PM10 : num 13.8 13.7 21.6 14.4 15.6 ...
## $ TCH : num 1.43 1.62 1.62 1.46 1.52 ...
## $ TOL : num 1.66 2.14 3.56 2.4 3.19 ...
## $ month : Date, format: "2011-01-01" "2011-01-01" ...
## $ week : Date, format: "2010-12-27" "2010-12-27" ...
## $ temp_avg : num 8.3 8.6 4.2 6.5 8.9 12.2 10.9 9.8 8.4 6.9 ...
## $ temp_max : num 13 13 9.4 8 10 15 13 13 11 8.3 ...
## $ temp_min : num 3 4 -1.6 4.1 6.3 8.9 8.7 7.6 6.6 5.2 ...
## $ precipitation : num 0 0 0 0 0 ...
## $ humidity : num 84 81 86 93 90 87 81 88 92 91 ...
## $ wind_avg_speed: num 5.2 5.4 3.5 6.3 10.4 15.7 15.6 14.3 6.5 7.4 ...
## $ temp_gap : num 10 9 11 3.9 3.7 6.1 4.3 5.4 4.4 3.1 ...
summary(pollution_daily_h)
## date NO2 SO2 O3
## Min. :2011-01-01 Min. : 7.819 Min. : 1.679 Min. : 3.979
## 1st Qu.:2012-07-01 1st Qu.: 26.170 1st Qu.: 3.757 1st Qu.: 30.088
## Median :2013-12-31 Median : 35.646 Median : 5.483 Median : 49.555
## Mean :2013-12-31 Mean : 38.828 Mean : 5.889 Mean : 47.866
## 3rd Qu.:2015-07-02 3rd Qu.: 48.122 3rd Qu.: 7.388 3rd Qu.: 65.058
## Max. :2016-12-31 Max. :105.077 Max. :17.129 Max. :110.287
## PM2.5 BEN CO EBE
## Min. : 2.625 Min. :0.1514 Min. :0.1496 Min. :0.1028
## 1st Qu.: 7.306 1st Qu.:0.4520 1st Qu.:0.2575 1st Qu.:0.3318
## Median :10.024 Median :0.6152 Median :0.3075 Median :0.6782
## Mean :11.112 Mean :0.7392 Mean :0.3574 Mean :0.6789
## 3rd Qu.:13.646 3rd Qu.:0.8911 3rd Qu.:0.4058 3rd Qu.:0.8651
## Max. :58.556 Max. :2.8132 Max. :1.1792 Max. :3.2252
## NMHC NO PM10 TCH
## Min. :0.04319 Min. : 2.351 Min. : 3.618 Min. :1.063
## 1st Qu.:0.15269 1st Qu.: 6.973 1st Qu.: 12.730 1st Qu.:1.327
## Median :0.18694 Median : 11.814 Median : 18.556 Median :1.394
## Mean :0.20500 Mean : 23.681 Mean : 20.805 Mean :1.421
## 3rd Qu.:0.23620 3rd Qu.: 26.178 3rd Qu.: 25.560 3rd Qu.:1.482
## Max. :0.66167 Max. :214.569 Max. :216.628 Max. :2.412
## TOL month week
## Min. : 0.2667 Min. :2011-01-01 Min. :2010-12-27
## 1st Qu.: 1.6969 1st Qu.:2012-07-01 1st Qu.:2012-06-30
## Median : 2.4893 Median :2013-12-16 Median :2013-12-30
## Mean : 3.0021 Mean :2013-12-16 Mean :2013-12-28
## 3rd Qu.: 3.6722 3rd Qu.:2015-07-01 3rd Qu.:2015-06-29
## Max. :15.6062 Max. :2016-12-01 Max. :2016-12-26
## temp_avg temp_max temp_min precipitation
## Min. :-0.50 Min. : 3.30 Min. :-7.000 Min. : 0.0000
## 1st Qu.: 8.40 1st Qu.:14.00 1st Qu.: 3.000 1st Qu.: 0.0000
## Median :14.60 Median :21.10 Median : 8.900 Median : 0.0000
## Mean :15.45 Mean :22.04 Mean : 8.755 Mean : 0.9173
## 3rd Qu.:22.70 3rd Qu.:30.10 3rd Qu.:14.600 3rd Qu.: 0.0000
## Max. :32.30 Max. :42.00 Max. :25.800 Max. :48.0100
## humidity wind_avg_speed temp_gap
## Min. :15.00 Min. : 2.00 Min. : 2.10
## 1st Qu.:36.00 1st Qu.: 6.30 1st Qu.: 9.50
## Median :55.00 Median : 8.70 Median :14.10
## Mean :55.04 Mean :10.15 Mean :13.29
## 3rd Qu.:73.00 3rd Qu.:12.60 3rd Qu.:17.00
## Max. :99.00 Max. :35.60 Max. :24.50
The data frame doesn’t contain any NA across its 2192 observations and 22 variables.
Evolution of the Variables Over Time
The charts below describe the evolution of each variable over time.

Dark Orange = Main Pollutants | Light Orange = Other Pollutants | Blue = Weather Parameters
Seasonal cycles suggests that the weather has an influence on the level of some pollutants.
Correlation Matrix
A correlation matrix is plotted to identify correlations between the variables:

Another view provides more information: 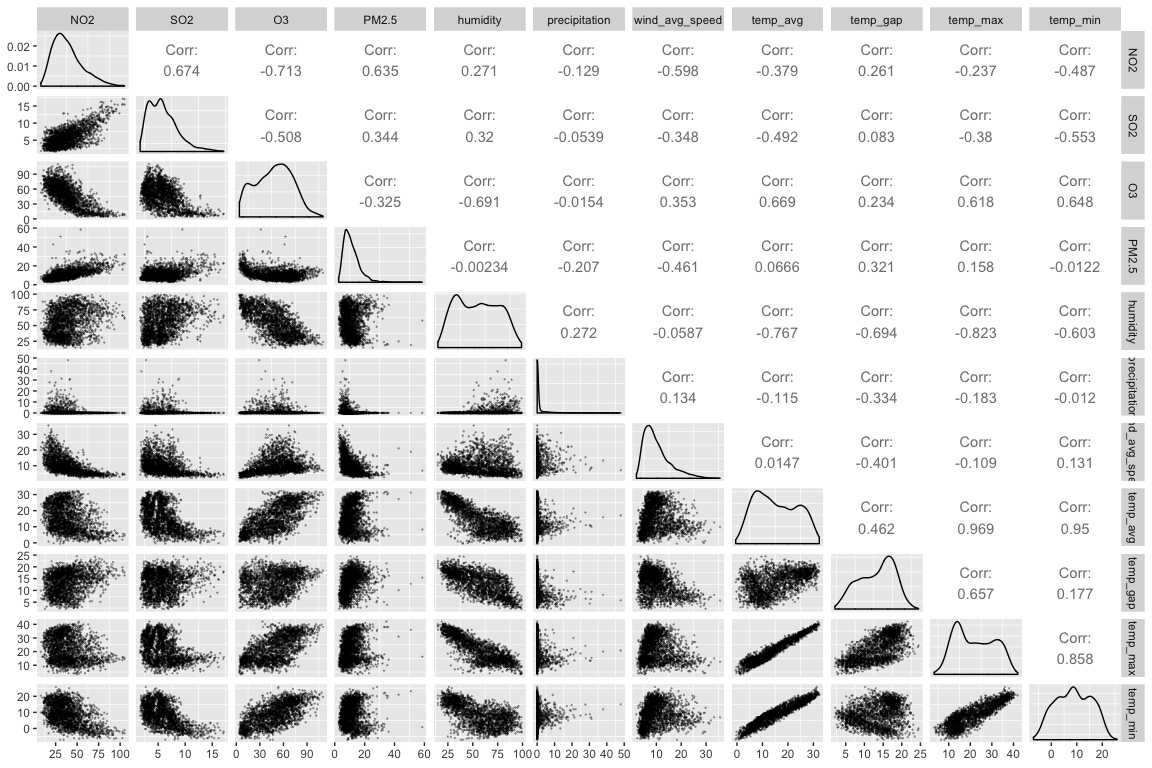
NO2 Model
A linear regression will be used to model the level of pollution in NO2. The data is split in train and test sets with the ratio 80|20.
set.seed(2018)
train.size <- 0.8
train.index <- sample.int(length(pollution_daily_h$NO2), round(length(pollution_daily_h$NO2) * train.size))
train.sample <- pollution_daily_h[train.index,]
test.sample <- pollution_daily_h[-train.index,]
The Train set has 1,754 rows and the Test set has 438 rows.
temp_min and temp_max are removed as correlated by definition with temp_avg. But the variable temp_gap is created to measure their influence on the model.
multi_model_NO2<-lm(NO2~.-month-week-date-temp_min-temp_max, data=train.sample)
lm_stats <- summary(multi_model_NO2)
print(lm_stats)
##
## Call:
## lm(formula = NO2 ~ . - month - week - date - temp_min - temp_max,
## data = train.sample)
##
## Residuals:
## Min 1Q Median 3Q Max
## -21.9368 -3.1235 0.0047 3.2157 21.3306
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.229319 3.113391 0.395 0.693003
## SO2 1.383111 0.077489 17.849 < 2e-16 ***
## O3 -0.149220 0.012032 -12.402 < 2e-16 ***
## PM2.5 0.255679 0.067646 3.780 0.000162 ***
## BEN 3.343452 1.165812 2.868 0.004182 **
## CO 45.100966 3.957109 11.397 < 2e-16 ***
## EBE 2.843966 0.553288 5.140 3.06e-07 ***
## NMHC 1.358368 1.981506 0.686 0.493105
## NO -0.201584 0.017344 -11.623 < 2e-16 ***
## PM10 -0.008354 0.024343 -0.343 0.731516
## TCH 11.070151 1.621469 6.827 1.19e-11 ***
## TOL 2.850827 0.214695 13.279 < 2e-16 ***
## temp_avg -0.122812 0.038525 -3.188 0.001459 **
## precipitation 0.178564 0.041030 4.352 1.43e-05 ***
## humidity -0.098214 0.015546 -6.318 3.36e-10 ***
## wind_avg_speed -0.323058 0.032912 -9.816 < 2e-16 ***
## temp_gap 0.304426 0.053019 5.742 1.10e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 5.096 on 1737 degrees of freedom
## Multiple R-squared: 0.9113, Adjusted R-squared: 0.9105
## F-statistic: 1116 on 16 and 1737 DF, p-value: < 2.2e-16
The R-square of the model is 0.9113 and the Adjusted R-squared is 0.9105, which means that the model is able to well explain NO2. Precisely, the predictors explain 91.1% of the variability in NO2.
The Mean Squared Error measures the mean of all of our errors squared. It describes the accuracy of a model. The MSE of this model is 5.0962.
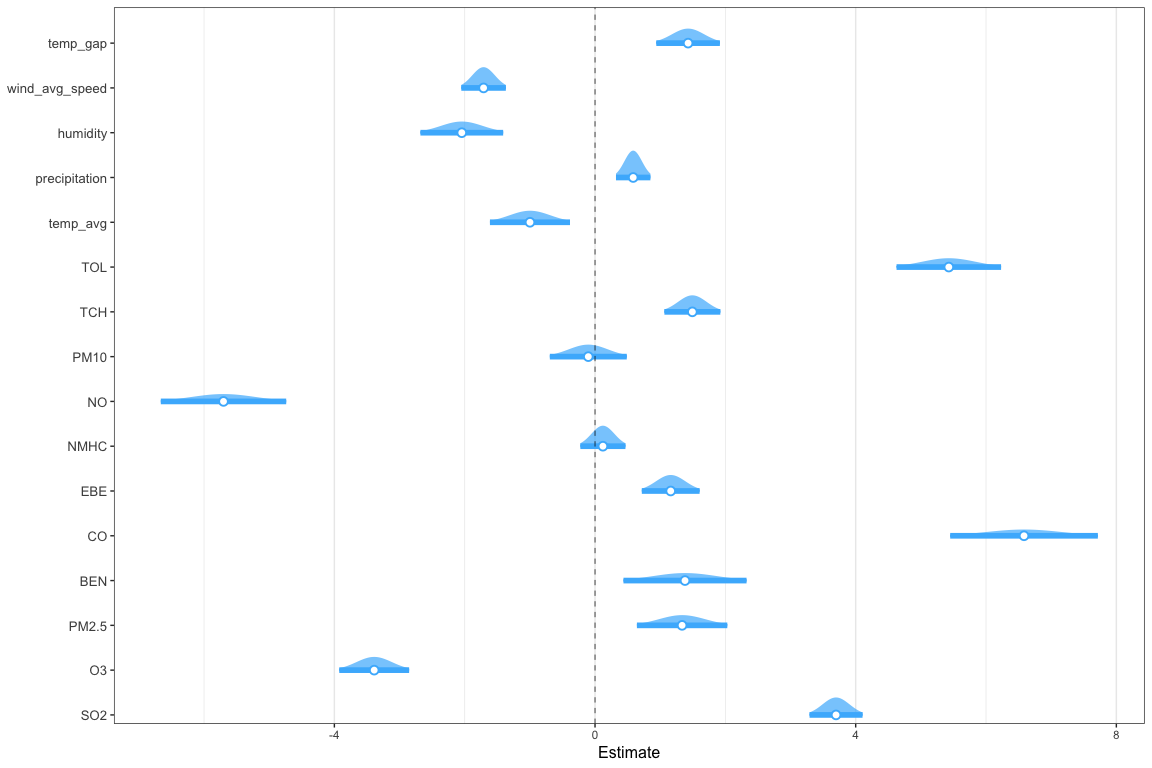
Another way to evaluate a model is looking at the confidence intervals of the coefficients. The estimates for each coefficient are not exact, so the confidence intervals define a range in which the actual values are, at a certain level of confidence:
For every change of one (1) unit in the SO2 level, one can be 95% confident that the level of NO2 will change between 1.23 and 1.54.
The confidence intervals can be plotted:
The residuals of the model can be checked using these plots:
resids_multi_NO2 <- multi_model_NO2$residuals
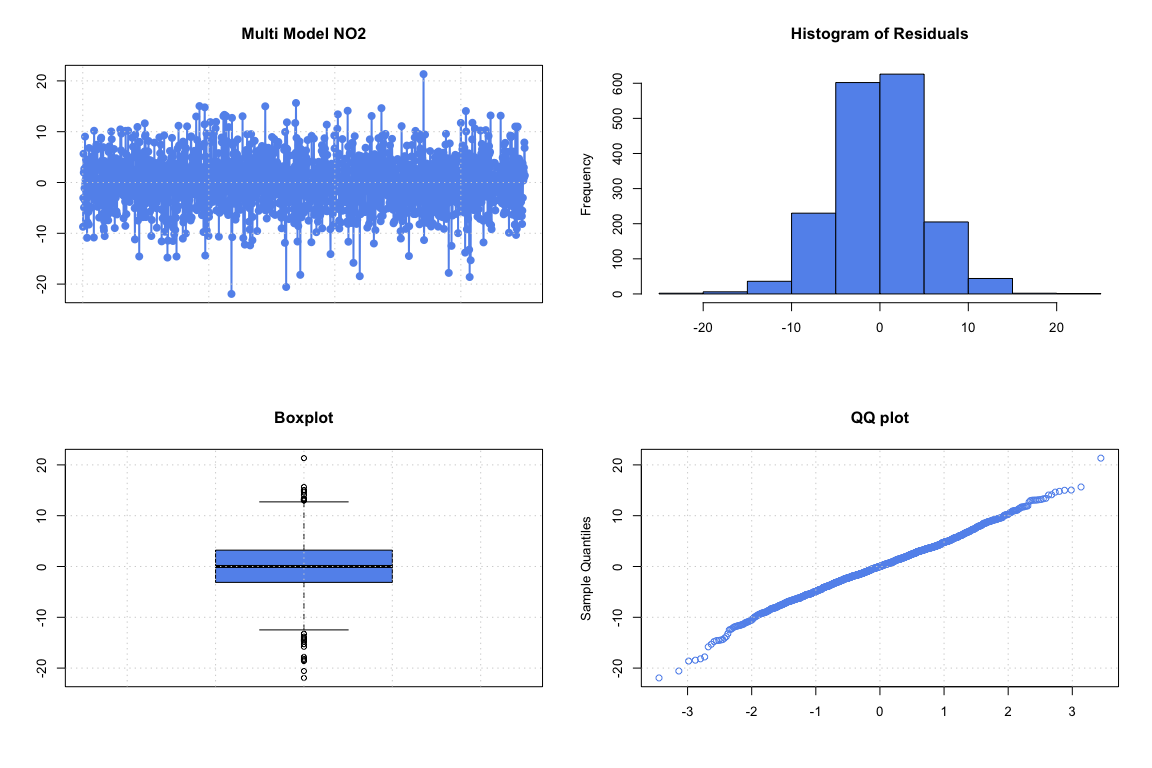
The residuals seem correct and validate the model. Significant variables can be found using a Stepwise Regression.
## Start: AIC=5729.65
## NO2 ~ (date + SO2 + O3 + PM2.5 + BEN + CO + EBE + NMHC + NO +
## PM10 + TCH + TOL + month + week + temp_avg + temp_max + temp_min +
## precipitation + humidity + wind_avg_speed + temp_gap) - month -
## week - date - temp_min - temp_max
##
## Df Sum of Sq RSS AIC
## - PM10 1 3.1 45114 5727.8
## - NMHC 1 12.2 45124 5728.1
## <none> 45111 5729.7
## - BEN 1 213.6 45325 5735.9
## - temp_avg 1 263.9 45375 5737.9
## - PM2.5 1 371.0 45482 5742.0
## - precipitation 1 491.9 45603 5746.7
## - EBE 1 686.2 45798 5754.1
## - temp_gap 1 856.2 45968 5760.6
## - humidity 1 1036.6 46148 5767.5
## - TCH 1 1210.5 46322 5774.1
## - wind_avg_speed 1 2502.4 47614 5822.3
## - CO 1 3373.7 48485 5854.2
## - NO 1 3508.4 48620 5859.0
## - O3 1 3994.5 49106 5876.5
## - TOL 1 4579.1 49691 5897.2
## - SO2 1 8274.0 53385 6023.0
##
## Step: AIC=5727.77
## NO2 ~ SO2 + O3 + PM2.5 + BEN + CO + EBE + NMHC + NO + TCH + TOL +
## temp_avg + precipitation + humidity + wind_avg_speed + temp_gap
##
## Df Sum of Sq RSS AIC
## - NMHC 1 11.8 45126 5726.2
## <none> 45114 5727.8
## + PM10 1 3.1 45111 5729.7
## - BEN 1 226.7 45341 5734.6
## - temp_avg 1 268.4 45383 5736.2
## - precipitation 1 489.2 45604 5744.7
## - EBE 1 684.2 45799 5752.2
## - temp_gap 1 860.1 45974 5758.9
## - PM2.5 1 1026.3 46141 5765.2
## - humidity 1 1067.1 46181 5766.8
## - TCH 1 1212.3 46327 5772.3
## - wind_avg_speed 1 2579.1 47693 5823.3
## - CO 1 3375.5 48490 5852.3
## - NO 1 3519.4 48634 5857.5
## - O3 1 4095.4 49210 5878.2
## - TOL 1 4582.1 49697 5895.4
## - SO2 1 8271.4 53386 6021.0
##
## Step: AIC=5726.23
## NO2 ~ SO2 + O3 + PM2.5 + BEN + CO + EBE + NO + TCH + TOL + temp_avg +
## precipitation + humidity + wind_avg_speed + temp_gap
##
## Df Sum of Sq RSS AIC
## <none> 45126 5726.2
## + NMHC 1 11.8 45114 5727.8
## + PM10 1 2.6 45124 5728.1
## - BEN 1 219.7 45346 5732.7
## - temp_avg 1 285.4 45412 5735.3
## - precipitation 1 493.7 45620 5743.3
## - EBE 1 674.1 45800 5750.2
## - temp_gap 1 863.8 45990 5757.5
## - PM2.5 1 1015.2 46141 5763.2
## - humidity 1 1089.1 46215 5766.1
## - TCH 1 2061.8 47188 5802.6
## - wind_avg_speed 1 2568.7 47695 5821.3
## - NO 1 3616.4 48743 5859.4
## - CO 1 3638.7 48765 5860.2
## - O3 1 4133.0 49259 5877.9
## - TOL 1 4701.8 49828 5898.1
## - SO2 1 8611.7 53738 6030.6
## Stepwise Model Path
## Analysis of Deviance Table
##
## Initial Model:
## NO2 ~ (date + SO2 + O3 + PM2.5 + BEN + CO + EBE + NMHC + NO +
## PM10 + TCH + TOL + month + week + temp_avg + temp_max + temp_min +
## precipitation + humidity + wind_avg_speed + temp_gap) - month -
## week - date - temp_min - temp_max
##
## Final Model:
## NO2 ~ SO2 + O3 + PM2.5 + BEN + CO + EBE + NO + TCH + TOL + temp_avg +
## precipitation + humidity + wind_avg_speed + temp_gap
##
##
## Step Df Deviance Resid. Df Resid. Dev AIC
## 1 1737 45111.36 5729.651
## 2 - PM10 1 3.058383 1738 45114.42 5727.770
## 3 - NMHC 1 11.762300 1739 45126.18 5726.227
These results indicate that the variables PM10 and NMHC can be removed. The resulting model is:
##
## Call:
## lm(formula = NO2 ~ . - month - week - date - temp_min - temp_max -
## PM10 - NMHC, data = train.sample)
##
## Residuals:
## Min 1Q Median 3Q Max
## -22.0494 -3.1185 0.0019 3.2071 21.4585
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.45196 2.92446 0.155 0.87720
## SO2 1.36999 0.07520 18.217 < 2e-16 ***
## O3 -0.14716 0.01166 -12.620 < 2e-16 ***
## PM2.5 0.23432 0.03746 6.255 5.00e-10 ***
## BEN 3.33921 1.14768 2.910 0.00367 **
## CO 45.69600 3.85895 11.842 < 2e-16 ***
## EBE 2.79371 0.54813 5.097 3.83e-07 ***
## NO -0.20239 0.01714 -11.805 < 2e-16 ***
## TCH 11.71564 1.31433 8.914 < 2e-16 ***
## TOL 2.86320 0.21271 13.461 < 2e-16 ***
## temp_avg -0.12662 0.03818 -3.317 0.00093 ***
## precipitation 0.17861 0.04095 4.362 1.37e-05 ***
## humidity -0.09773 0.01509 -6.478 1.20e-10 ***
## wind_avg_speed -0.32267 0.03243 -9.949 < 2e-16 ***
## temp_gap 0.30558 0.05297 5.769 9.40e-09 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 5.094 on 1739 degrees of freedom
## Multiple R-squared: 0.9113, Adjusted R-squared: 0.9106
## F-statistic: 1276 on 14 and 1739 DF, p-value: < 2.2e-16
The R-squared value of 0.9113 is consistent with the initial model.
Multicollinearity can be treated with the VIF Method (Variance Inflation Factors). As a general rule, if the VIF value is larger than 5, the multicollinearity is assumed to be high.
For each variable of the model:
- the VIF values are calculated,
- the variable with the largest value is removed,
- the model is re-run the explanatory variables having a VIF value below 5.
The current VIF values are:
## SO2 O3 PM2.5 BEN CO
## 2.731960 4.737638 2.589103 15.175970 21.433198
## EBE NO TCH TOL temp_avg
## 3.384605 15.882579 2.124477 11.084651 6.505867
## precipitation humidity wind_avg_speed temp_gap
## 1.218099 6.663257 1.991433 4.170651
The VIF values resulting from the procedure are:
## SO2 O3 PM2.5 EBE TCH
## 1.958788 3.351606 2.049109 1.829030 1.781684
## temp_avg precipitation wind_avg_speed temp_gap
## 3.056575 1.154820 1.743077 2.058636
After removing multicollinear variables, the Final Model is:
## NO2 ~ SO2 + O3 + PM2.5 + EBE + TCH + temp_avg + precipitation +
## wind_avg_speed + temp_gap
##
## Call:
## lm(formula = NO2 ~ SO2 + O3 + PM2.5 + EBE + TCH + temp_avg +
## precipitation + wind_avg_speed + temp_gap, data = train.sample)
##
## Residuals:
## Min 1Q Median 3Q Max
## -21.9354 -4.1242 0.1759 3.9862 18.8808
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -1.34165 2.32129 -0.578 0.5634
## SO2 1.70825 0.07619 22.421 < 2e-16 ***
## O3 -0.23205 0.01174 -19.774 < 2e-16 ***
## PM2.5 0.56677 0.03988 14.213 < 2e-16 ***
## EBE 8.34368 0.48211 17.307 < 2e-16 ***
## TCH 17.17983 1.44013 11.929 < 2e-16 ***
## temp_avg -0.13117 0.03131 -4.189 2.94e-05 ***
## precipitation 0.14279 0.04770 2.993 0.0028 **
## wind_avg_speed -0.35111 0.03630 -9.672 < 2e-16 ***
## temp_gap 0.77269 0.04452 17.355 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 6.095 on 1744 degrees of freedom
## Multiple R-squared: 0.8727, Adjusted R-squared: 0.872
## F-statistic: 1328 on 9 and 1744 DF, p-value: < 2.2e-16
A 10-Fold Cross Validation can confirm the accuracy of the models:
## [1] "Initial Model:"
## Linear Regression
##
## 1754 samples
## 21 predictor
##
## Pre-processing: centered (16), scaled (16)
## Resampling: Cross-Validated (10 fold)
## Summary of sample sizes: 1578, 1579, 1579, 1579, 1578, 1579, ...
## Resampling results:
##
## RMSE Rsquared MAE
## 5.13777 0.9079525 3.97799
##
## Tuning parameter 'intercept' was held constant at a value of TRUE
## [1] "Final Model:"
## Linear Regression
##
## 1754 samples
## 9 predictor
##
## Pre-processing: centered (9), scaled (9)
## Resampling: Cross-Validated (10 fold)
## Summary of sample sizes: 1578, 1578, 1579, 1579, 1578, 1578, ...
## Resampling results:
##
## RMSE Rsquared MAE
## 6.112716 0.8716267 4.866125
##
## Tuning parameter 'intercept' was held constant at a value of TRUE
The predictions of both models can be compared.
test.sample$NO2_predicted_model_final <- predict(multi_model_NO2_final,test.sample)
test.sample$NO2_predicted_model_0 <- predict(multi_model_NO2_0,test.sample)
Depending on the prediction point, the final model can be better or worse than the initial model. Below table displays some examples of the prediction points (predictions rows 80-90):
## NO2 NO2_predicted_model_0 NO2_predicted_model_final
## 428 32.90278 33.47543 38.52169
## 436 41.62434 45.47223 48.86567
## 438 66.43728 62.02274 58.57356
## 441 37.94618 36.13524 35.07174
## 443 20.01215 21.83412 21.85867
## 447 58.33043 54.81361 52.00605
## 464 35.96528 37.69067 40.09754
## 470 18.65799 15.89007 17.62860
## 482 28.10417 21.46581 22.55336
## 489 45.54340 37.47732 37.65892
## 490 31.50521 22.42451 23.59314
The accuracy of each prediction point can be understood by comparing their values with the actual values:
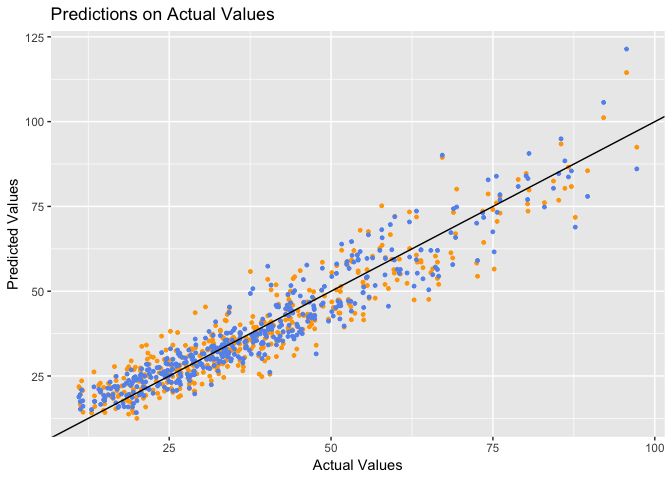
Blue = Initial Model | Orange = Final Model
The models predictions can be compared statistically using an One-Way Analysis of Variance (ANOVA) and a plot of the coefficients confidence intervals:
## Analysis of Variance Table
##
## Model 1: NO2 ~ (date + SO2 + O3 + PM2.5 + BEN + CO + EBE + NMHC + NO +
## PM10 + TCH + TOL + month + week + temp_avg + temp_max + temp_min +
## precipitation + humidity + wind_avg_speed + temp_gap) - month -
## week - date - temp_min - temp_max - PM10 - NMHC
## Model 2: NO2 ~ SO2 + O3 + PM2.5 + EBE + TCH + temp_avg + precipitation +
## wind_avg_speed + temp_gap
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 1739 45126
## 2 1744 64788 -5 -19661 151.54 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
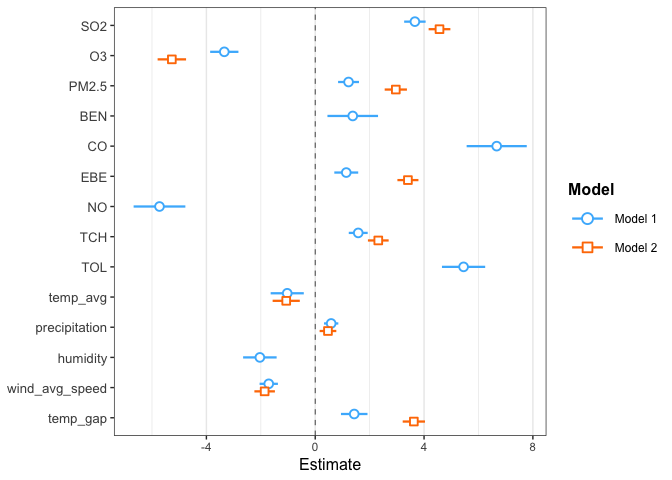
The low p-value returned by the ANOVA indicates that the Final model is significantly better than the Initial Model. The plot gives an indication on how each variable influences the predictions, with a 95% confidence interval.
As a conclusion, the Final Model provides a good way to predict the NO2 pollution level based on 9 pollutants and 5 weather parameters.
